

TopoTools consists of a generic middleware script layer that makes access to the topology related data stored in VMD more convenient than the existing very basic … gemeindeverein stettfurt It is meant to be a complementary tool to psfgen, which is very much optimized for building topologies for biomolecules. WebTopoTools is a VMD plugin for manipulating topology information. How could I select a range of ATOMS by using VMD Program? Vmd representation material VMD-L Mailing List WebControl panel → Graphics → Representation → select “Drawing method” as “cpk” or “point” or line mol representation CPK
Vmd tutorial files how to#
How to visualize protein-ligand complex MD run using VMD It may be used to … gimp bearbeitungsprogramm
Vmd tutorial files software#
(and you are showing the bonds in your VMD representation), the software will connect the O in one side, with the H that just appeared in the other side.

structure 2 gemeindeverfassungsrecht bayern For example, possible structure commands include. The "structure" command takes the following parameters: molecule id, atom selection, structure type. Yes, you can, with the VMD command "structure" which is accessible through the scripting interface.
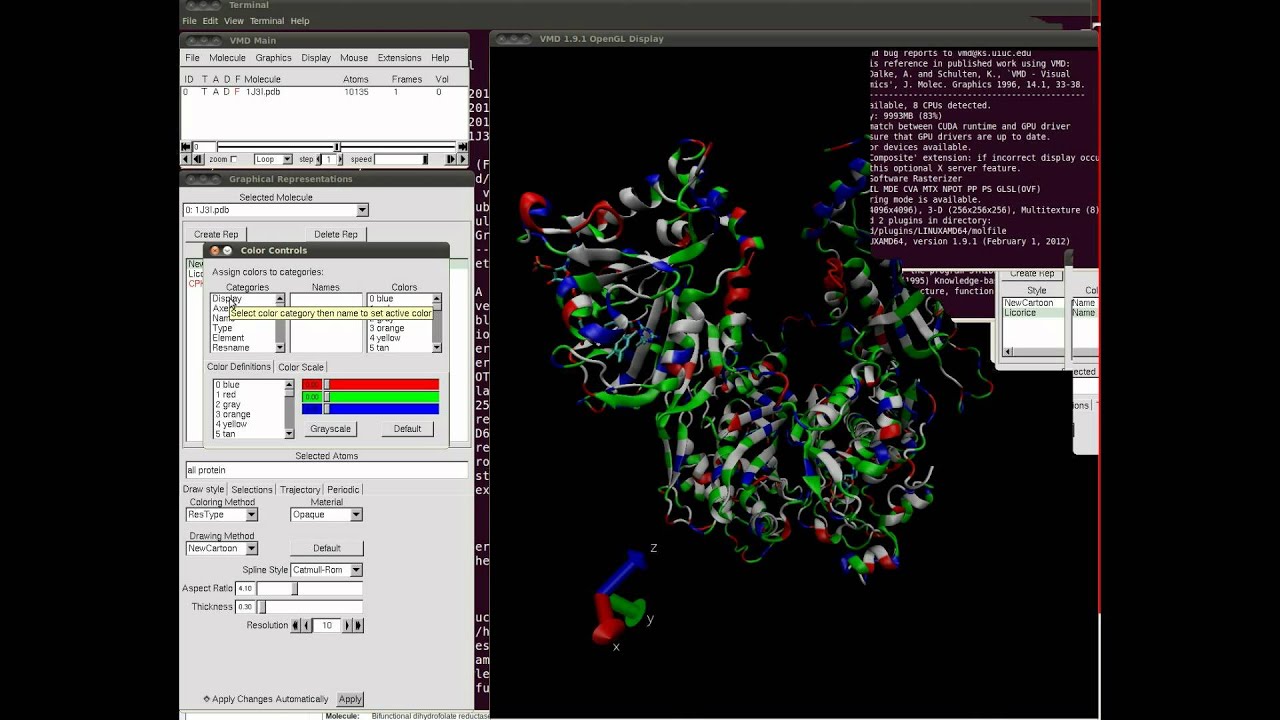
Molecular Representations in VMD - University of Illinois …


 0 kommentar(er)
0 kommentar(er)
